

When sequences in the source databases change, these changes are tracked by UniParc and history of all changes is archived. National Biomedical Research Foundation. It combines information extracted from scientific literature and biocurator -evaluated computational analysis.
- The mission of UniProt is to provide the scientific community with a comprehensive, high quality and freely accessible resource of protein sequence and functional information. Annotation arising from the scientific literature includes, but is not limited to:
- What is manual annotation? Further information How to cite UniProt Submissions and updates to UniProtKB UniProtKB entry view manual Printable Quick Guide to UniProtKB.
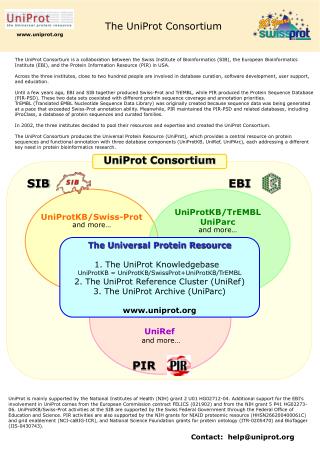
- The UniProt consortium maintains the UniProt KnowledgeBase UniProtKB and several supplementary databases including the UniProt Reference Clusters UniRef and the UniProt Archive UniParc.
- Author information 1 Swiss Institute of Bioinformatics, Centre Medical Universitaire, Geneva, Switzerland.
- TrEMBL 88,, Automatically annotated and not reviewed.
- In eukaryotes, the most widespread method is for two reproductive cells of the opposite sex to meet and fuse.
Interaction Help About Wikipedia Community portal Recent changes Contact page. Select item s and click on "Add to basket" to create your own collection here entries max. How do we manually annotate a UniProtKB entry?
Your basket is currently empty. By using this site, you agree to the Terms of Use and Privacy Policy.
UniProt
Text is available under the Creative Commons Attribution-ShareAlike License ; additional terms may apply. SIB, located in Geneva, Switzerland, maintains the ExPASy Expert Protein Analysis System servers that are a central resource for proteomics tools and databases.
Standard operating procedure SOP for UniProt manual curation. How to cite this comment: Align BLAST Map Ids Download Full View Remove Clear. Select item s and click on "Add to basket" to create your own collection here entries max.
UniProt - Wikipedia
By clicking or navigating the site, you agree to allow our collection of information on and off Facebook through cookies. Relevant publications are identified by searching databases such as PubMed.
UniProtKB/Swiss-Prot.
UniProt is comprised of four components, each optimised for different uses: In eukaryotes, the most widespread method is for two reproductive cells of the opposite sex to meet and fuse. PubMed US National Library of Medicine National Institutes of Health.
UniRef sequences are clustered using the CD-HIT algorithm to build UniRef90 and UniRef Articles containing potentially dated statements from March All articles containing potentially dated statements.
It contains a large amount of information about the biological function of proteins derived from the research literature. Homology BLAST Basic Local Alignment Search Tool BLAST Stand-alone BLAST Link BLink Conserved Domain Database CDD Conserved Domain Search Service CD Search Genome ProtMap HomoloGene Protein Clusters All Homology Resources Download Centre Release Statistics QuickGO Posters.
TrEMBL 88,, Automatically annotated and not reviewed. Custom flat file, FASTA , GFF , RDF , XML.
Swiss-Prot , Manually annotated and reviewed. Rules for automatic annotation generated by database curators and computational algorithms. Clustering sequences significantly reduces database size, enabling faster sequence searches. UniProt is the Uni versal Prot ein resource, a central repository of protein data created by combining the Swiss-Prot, TrEMBL and PIR-PSD databases.
Messenger Facebook Lite People Places Games Locations Celebrities Marketplace Groups Recipes Moments Instagram About Create Ad Create Page Developers Careers Privacy Cookies Ad Choices Terms Settings Activity Log.
Retrieved from " https: The UniProt consortium maintains the UniProt KnowledgeBase UniProtKB and several supplementary databases including the UniProt Reference Clusters UniRef and the UniProt Archive UniParc. The consortium members pooled their overlapping resources and expertise, and launched UniProt in December



[MYCB(RAMBLER)FREETEXT-1-2